Welcome to AcrTransAct, an advanced web application powered by a transformer-based Deep Neural Network
designed to predict the likelihood of Acr-mediated CRISPR-Cas inhibition.
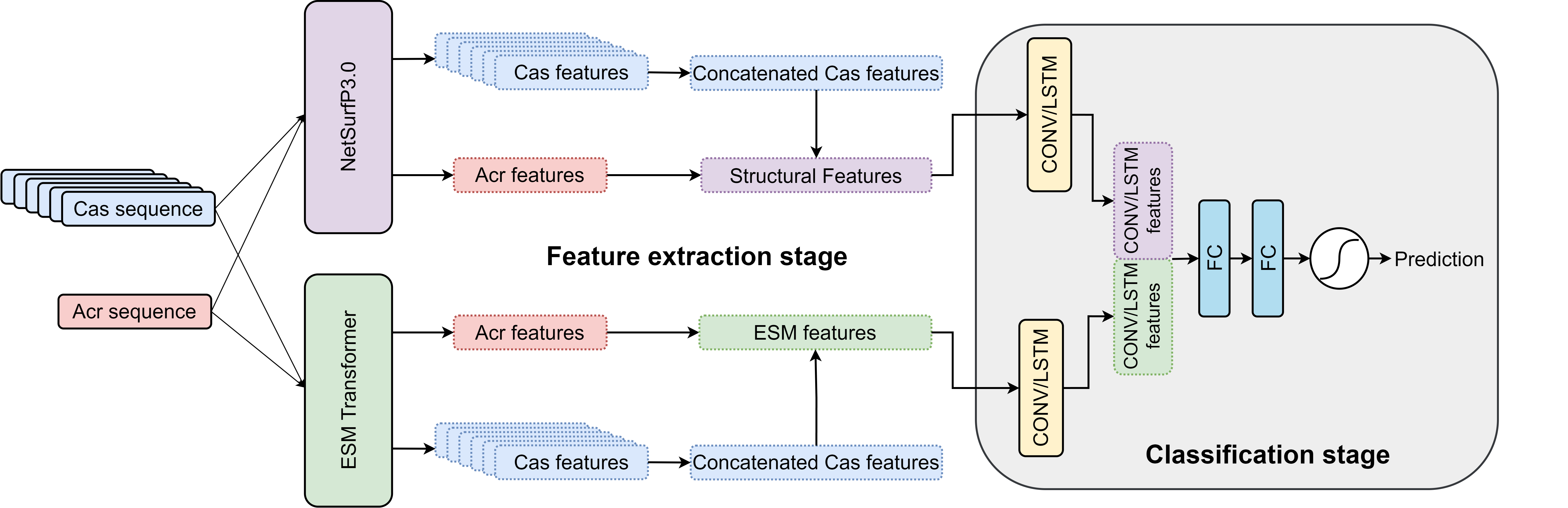
Our model consists of two main components:
1. Feature Extraction Module: This module incorporates a pre-trained Evolutionary Scale Modeling (ESM)
protein transformer and the NetSurfP-3.0 secondary structure prediction system. This module efficiently
extracts meaningful features from input protein sequences, capturing essential information such as
secondary structure.
2. Classification Module: Comprising either a CNN or LSTM network, this module utilizes the extracted
features from the previous module to classify whether a given Acr protein is likely to inhibit the input
CRISPR-Cas system.
Our model has been meticulously trained on an inhibition dataset of 227 experiments sourced from two
Acr databases, AcrHub and Anti-CRISPRdb, and various published works. The dataset encompasses verified
interactions of Acr with CRISPR-Cas systems for subtypes I-C, I-E, and I-F.
When you input a putative Acr protein sequence, it undergoes processing through the feature extraction
module. The ESM protein transformer analyzes the sequence and derives a set of essential features, and
the NetSurfp-3.0 network predicts Q3, ASA, RSA, and disorder for each residue. These extracted features
are then passed to the classification module, where they are processed by CNN/LSTM layers and passed to
fully connected layers. The classification module generates a probability of the input Acr sequence
inhibiting the input CRISPR-Cas system.
To ensure the model's reliability and effectiveness, we have meticulously divided our data into
training, validation, and test sets. We perform a thorough hyperparameter search on the
training-validation set to optimize model performance, and then rigorously test the top models on the
unseen test set. Our results showcase remarkable accuracy and F1 scores ranging from 0.93 to 0.95,
reflecting the exceptional predictive capabilities of AcrTransAct.
Model Architecture:
Our code and data are available at
Github.com/USask-BINFO/AcrTransAct
Cite our paper:
@inproceedings{hasani2023acrtransact,
author = {Moein Hasani and Chantel N. Trost and Nolen Timmerman and Lingling Jin},
title = {AcrTransAct: Pre-trained Protein Transformer Models for the Detection of Type I Anti-CRISPR Activities},
booktitle = {Proceedings of The 14th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics (ACM-BCB)},
year = {2023},
publisher = {ACM},
address = {Houston, TX, USA},
pages = {6},
}